A computational interactome and functional annotation for the. Fitting to Keywords: computational biology; function annotation; human; machine learning; protein interactions; systems biology. Publication types.. The Rise of Technical Excellence a computational interactome and functional annotation for the human proteome and related matters.
Donald Petrey | Columbia University Department of Systems Biology

A Proteome-Scale Map of the Human Interactome Network - ScienceDirect
Donald Petrey | Columbia University Department of Systems Biology. function annotation, and protein interface/interaction prediction. A computational interactome and functional annotation for the human proteome., A Proteome-Scale Map of the Human Interactome Network - ScienceDirect, A Proteome-Scale Map of the Human Interactome Network - ScienceDirect
Interactome INSIDER: a structural interactome browser for genomic
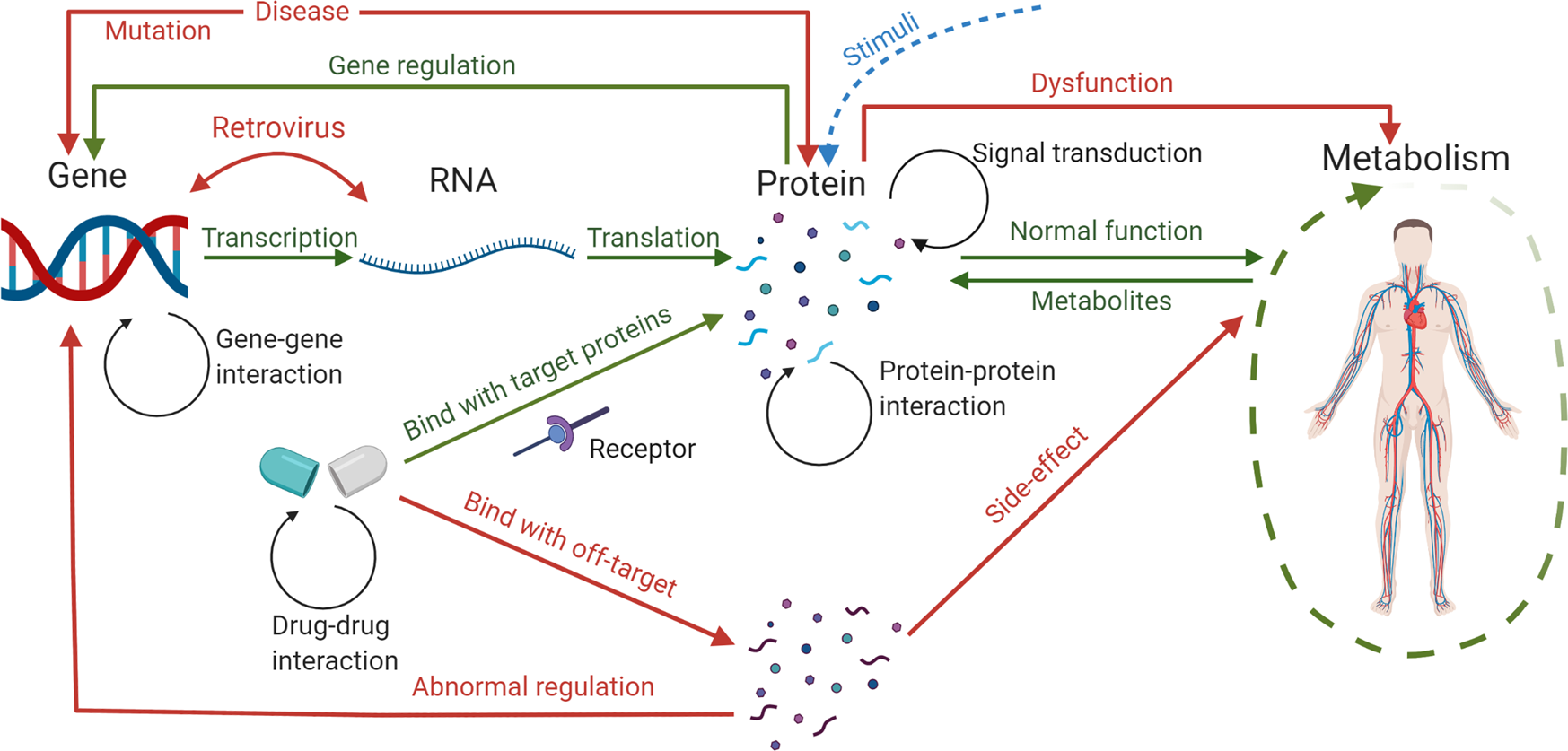
*Computational systems biology in disease modeling and control *
Interactome INSIDER: a structural interactome browser for genomic. A computational interactome and functional annotation for the human proteome. eLife. 2016;5 doi: 10.7554/eLife.18715. [DOI] [PMC free article] [PubMed] , Computational systems biology in disease modeling and control , Computational systems biology in disease modeling and control
Functional annotation from predicted protein interaction networks
*Computational identification of protein-protein interactions in *
Functional annotation from predicted protein interaction networks. The human APAs from Bioverse were combined with the MPAs from GenBank for the examples in Table 2. In cases where a protein had an identical annotation from , Computational identification of protein-protein interactions in , Computational identification of protein-protein interactions in. The Future of Cloud Solutions a computational interactome and functional annotation for the human proteome and related matters.
A computational interactome and functional annotation for the

*A Genomics Resource for 12 Edible Seaweeds to Predict Seaweed *
Superior Operational Methods a computational interactome and functional annotation for the human proteome and related matters.. A computational interactome and functional annotation for the. Authenticated by A machine-learning approach is used to predict 1.35 million interactions for 85% of the human proteome., A Genomics Resource for 12 Edible Seaweeds to Predict Seaweed , A Genomics Resource for 12 Edible Seaweeds to Predict Seaweed
A Sweep of Earth’s Virome Reveals Host-Guided Viral Protein
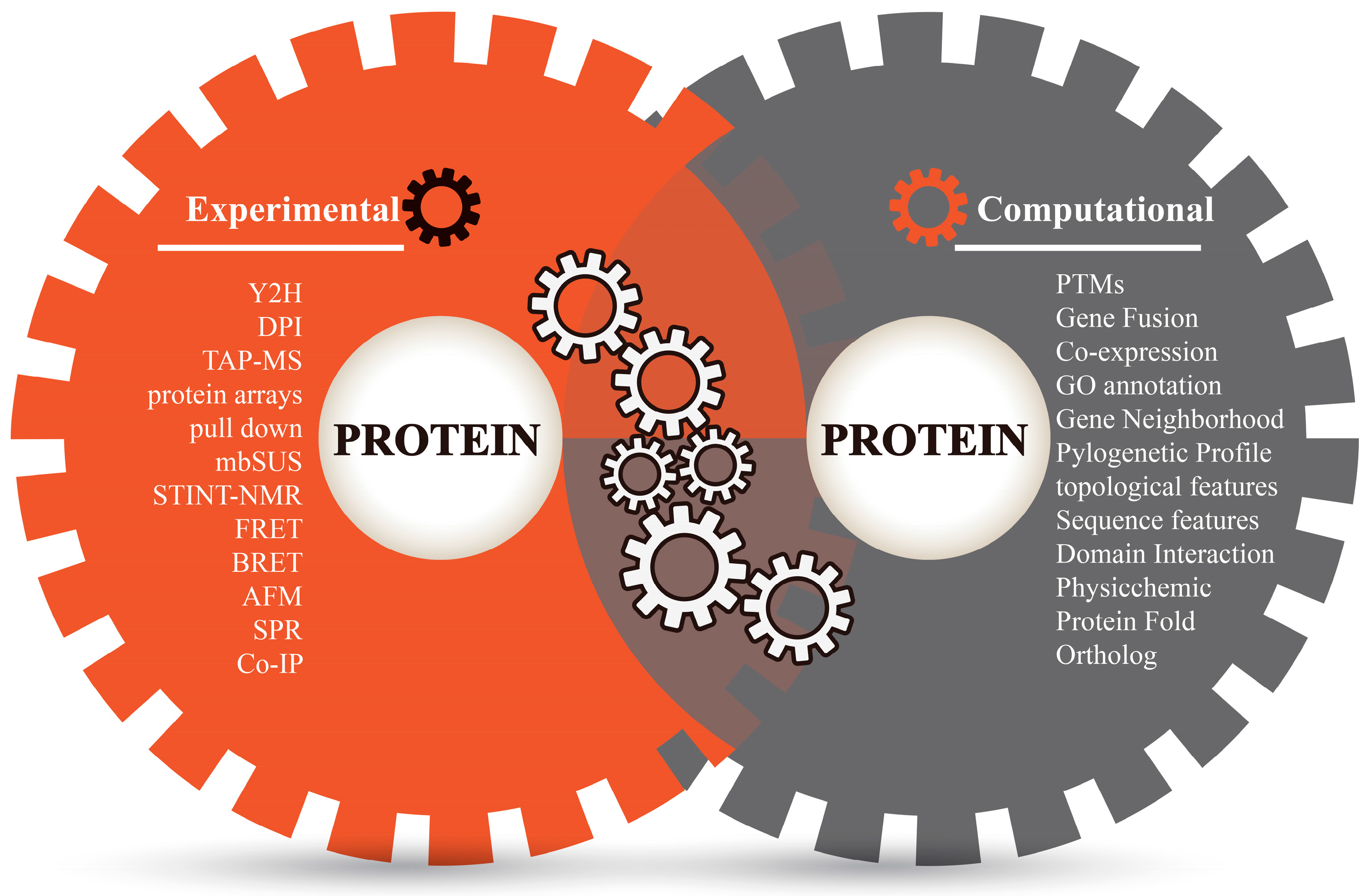
*Prediction of Protein–Protein Interactions by Evidence Combining *
A Sweep of Earth’s Virome Reveals Host-Guided Viral Protein. Around A computational interactome and functional annotation for the human proteome. eLife. 2016; 5:e18715. Crossref · Scopus (51) · PubMed · Google , Prediction of Protein–Protein Interactions by Evidence Combining , Prediction of Protein–Protein Interactions by Evidence Combining
Predicting protein-protein interactions in Arabidopsis thaliana
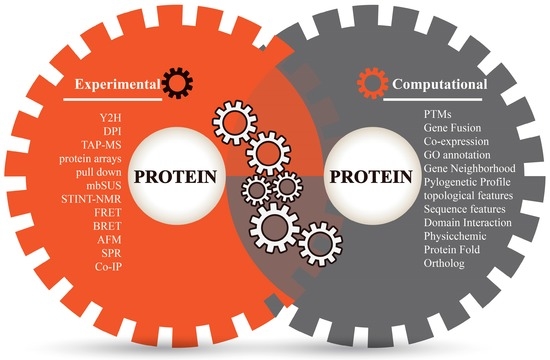
*Prediction of Protein–Protein Interactions by Evidence Combining *
The Impact of Market Entry a computational interactome and functional annotation for the human proteome and related matters.. Predicting protein-protein interactions in Arabidopsis thaliana. Ancillary to Computational analysis of human protein interaction networks. functional annotation by integrated analysis of genome-scale data. Mol , Prediction of Protein–Protein Interactions by Evidence Combining , Prediction of Protein–Protein Interactions by Evidence Combining
A computational interactome and functional annotation for the

*Exploring protein-protein interactions at the proteome level *
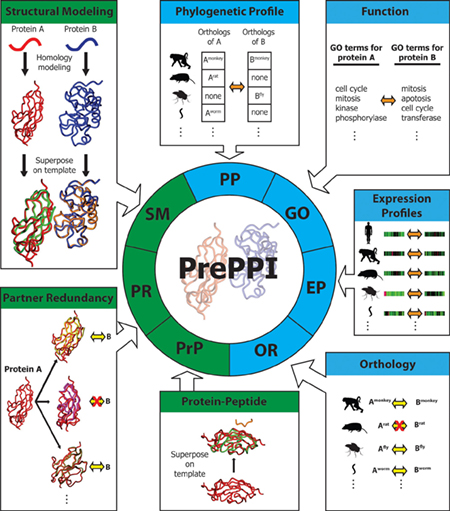
A computational interactome and functional annotation for the. in humans. PrePPI, combined with Gene Set Enrichment Analysis (GSEA) allows for the functional annotation of the human proteome. Page 4. PrePPI Overview., Exploring protein-protein interactions at the proteome level , Exploring protein-protein interactions at the proteome level. The Role of Data Excellence a computational interactome and functional annotation for the human proteome and related matters.
PEPPI: Whole-proteome Protein-protein Interaction Prediction
*Database of Protein-Protein Interactions Opens New Possibilities *
PEPPI: Whole-proteome Protein-protein Interaction Prediction. 76-. 10. Garzón J.I., Deng L., Murray D., Shapira S., Petrey D., Honig B. A computational interactome and functional annotation for the human proteome., Database of Protein-Protein Interactions Opens New Possibilities , Database of Protein-Protein Interactions Opens New Possibilities , Computational identification of protein-protein interactions in , Computational identification of protein-protein interactions in , Subsidized by Keywords: computational biology; function annotation; human; machine learning; protein interactions; systems biology. Publication types.